About
Hey there! I'm currently a third-year Ph.D. student in Computer Science and Technology at Tongji University, supervised by Prof. Guang Chen, the head of the Generalist Embodied AI Lab. My research focuses on AI4Science, with specific interests in (1) drug discovery, (2) generative models, and (3) reinforcement learning.
📣 News
- 2025.08 Our work (SWITCH) on spatial multi-omics is accepted by Nature Computational Science (NCS)!
- 2025.02 Our work (RCP-Bench) on collaborative perception is accepted by CVPR 2025!
- 2024.12 We won the championship (1/226) in The Second Global AI Drug Development Algorithm Competition!
- 2024.11 Our work (AMG) on 3D structure-based molecular generation is accepted by Briefings in Bioinformatics!
- 2022.09 Our work on medical image segmentation is accepted by IEEE JBHI (IF: 7.7)!
- 2022.07 Our work (Molormer) on drug-drug interaction prediction is accepted by Briefings in Bioinformatics (IF: 13.994)!
- 2021.12 Our work on drug-drug interaction prediction is accepted by Briefings in Bioinformatics (IF: 13.994)!
📝 Publications
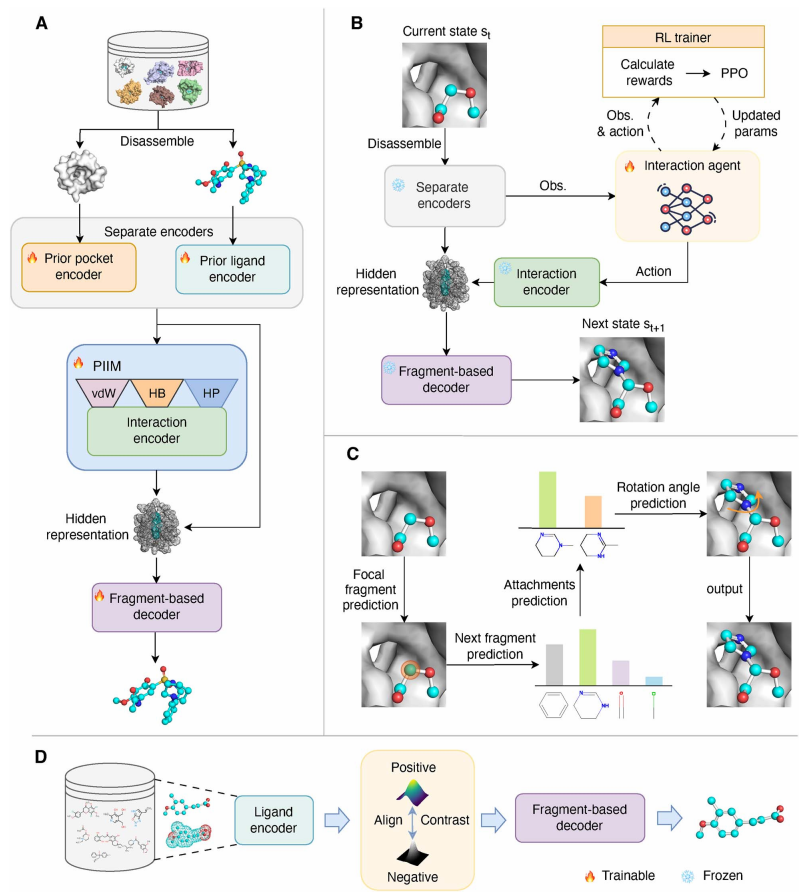
Deep reinforcement learning as an interaction agent to steer fragment-based 3D molecular generation for protein pockets
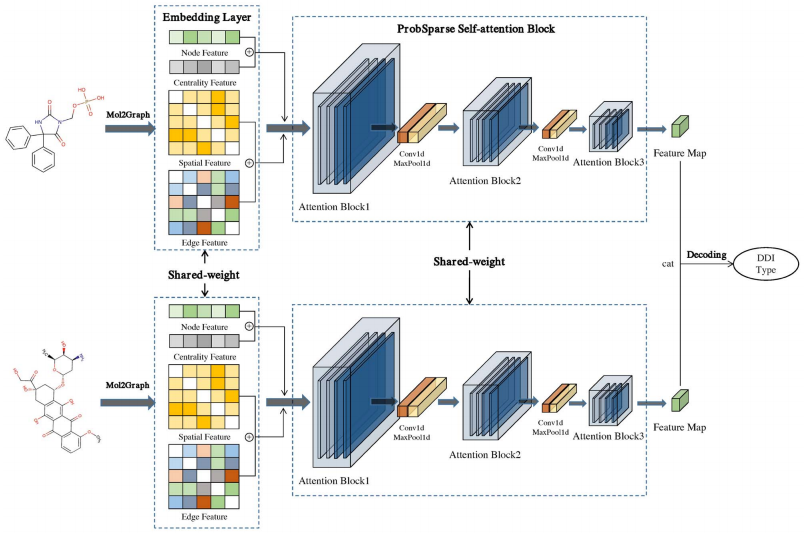
Molormer: a lightweight self-attention-based method focused on spatial structure of molecular graph for drug-drug interactions prediction
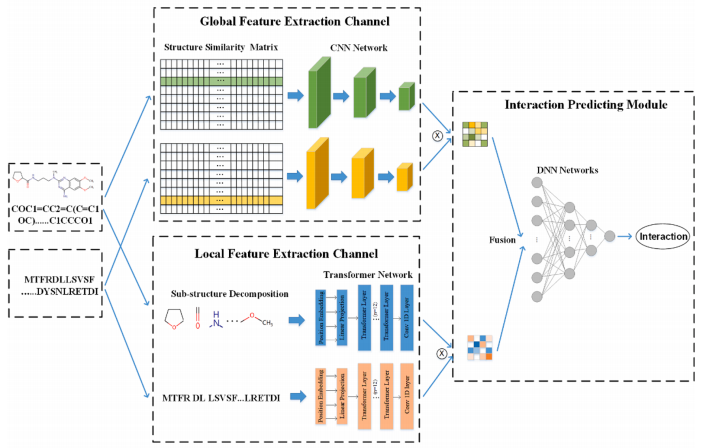
DeepFusion: A Deep Learning Based Multi-Scale Feature Fusion Method for Predicting Drug-Target Interactions
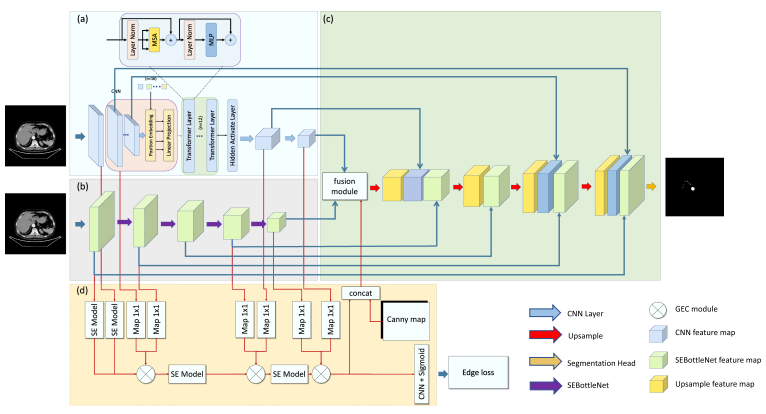
TransFusionNet: Semantic and Spatial Features Fusion Framework for Liver Tumor and Vessel Segmentation Under JetsonTX2
💼 Experience
Research Intern
XtalPi (Ailux & 抗体IDD部)Conducting research on AI antibody design, focusing on developing computational methods for antibody structure prediction and optimization.
🏆 Honors and Awards
- 2025.10 Special Prize and Most Popular Award, 19th Challenge Cup AI+ Special Competition
- 2024.12 First Prize in the 2nd Global AI Drug Development Algorithm Competition (1/226)
- 2023.04 Outstanding Graduate of Shandong Province
- 2022.12 National scholarship for Postgraduates
🌐 Academic Service
I serve as a reviewer for Briefings in Bioinformatics, JBHI, Journal of Cheminformatics, BioData Mining, and BMC Bioinformatics.
